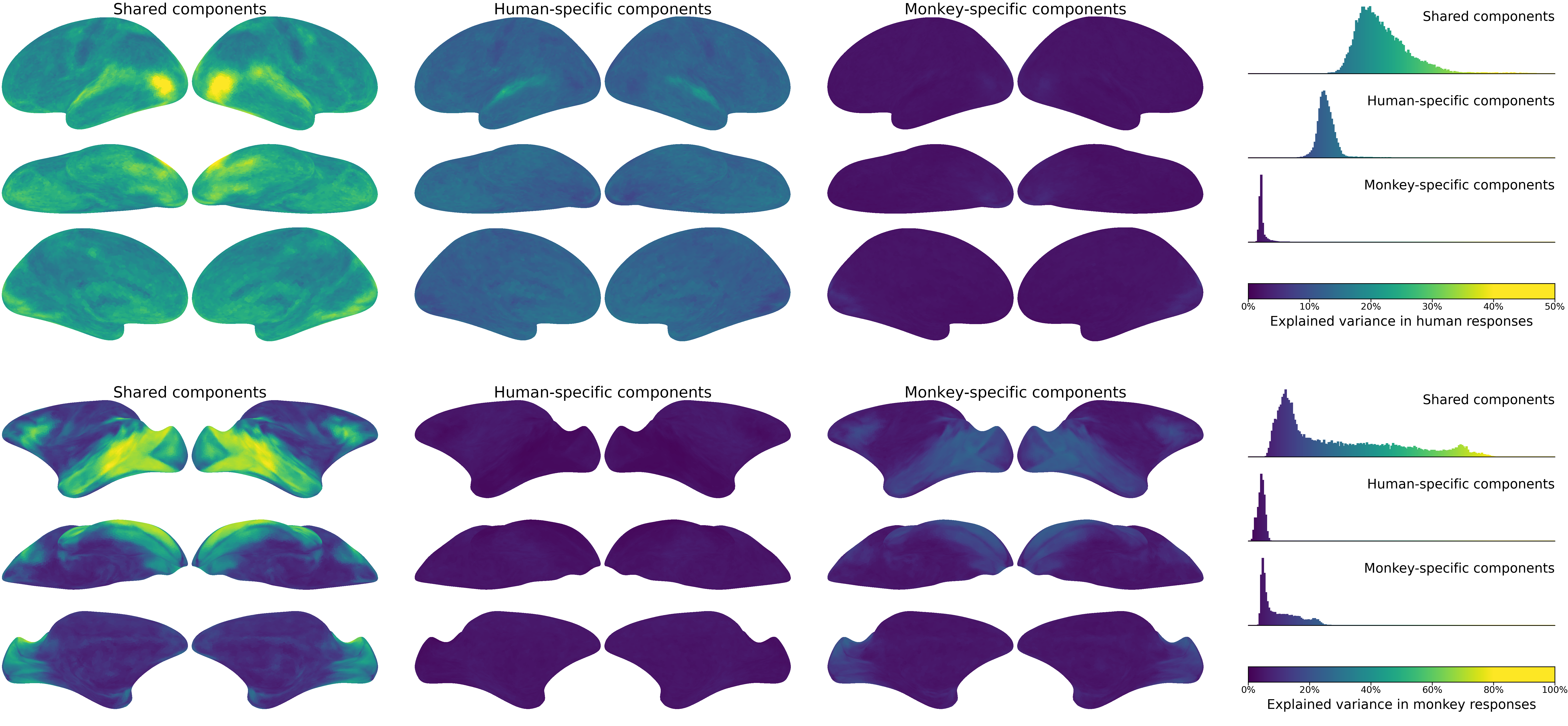
Comparisons across species and systems#
The same function can be implemented in different ways [Haxby et al., 2020]. To understand the general principles of brain functional organization, we examine how the same function is instantiated in different species (e.g., humans and non-human primates) and systems (e.g., brains and artificial neural networks).
Modeling human brains using deep neural networks#
Are deep neural networks (DNNs) good models of the human face processing system? To address this question, we systematically compared how 707 face video clips are represented in human brains and in DNNs [Jiahui et al., 2023]. We found systematic and meaningful correlations between human brain representations and DNN representations throughout the face processing network. However, DNN representations only explain a portion of variance in human brain representations, suggesting the existence of differences between biological and artificial face processing systems.
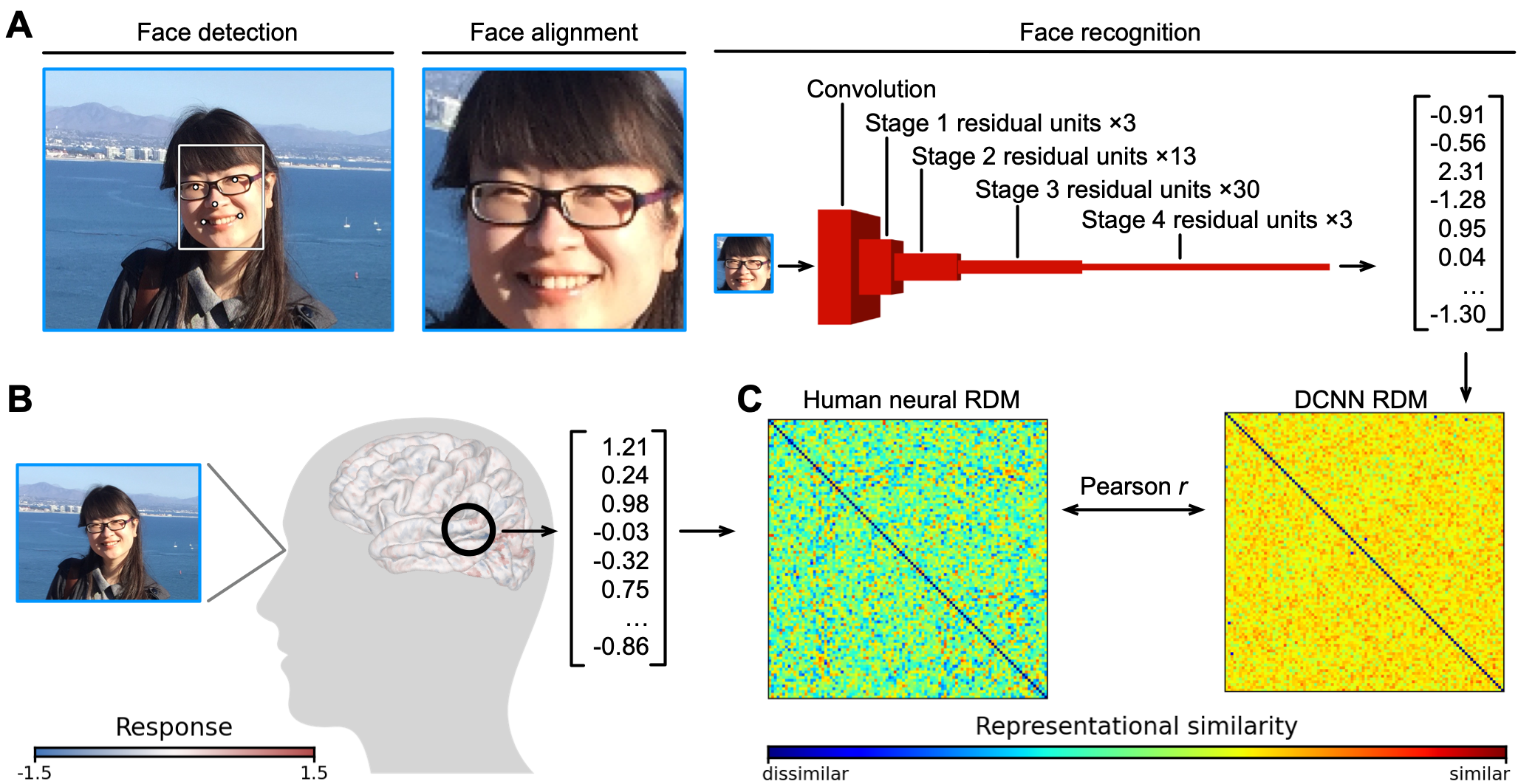
Fig 1. Comparison between human brain and DNN representations. The representation of a face is characterized as a vector in each brain region or DNN layer. Based on the similarities between these vectors, we constructed representational dissimilarity matrices (RDMs) for each brain region and DNN layer and systematically compared them.
References#
James V Haxby, J Swaroop Guntupalli, Samuel A Nastase, and Ma Feilong. Hyperalignment: modeling shared information encoded in idiosyncratic cortical topographies. elife, 9:e56601, 2020. doi:https://doi.org/10.7554/eLife.56601.
Guo Jiahui, Ma Feilong, Matteo Visconti di Oleggio Castello, Samuel A Nastase, James V Haxby, and M Ida Gobbini. Modeling naturalistic face processing in humans with deep convolutional neural networks. Proceedings of the National Academy of Sciences, 120(43):e2304085120, 2023. doi:https://doi.org/10.1073/pnas.2304085120.