Brain differences and their correlates#
A key focus of our lab is understanding individual differences in the human brain, including:
how brains differ from each other;
how these differences develop across the lifespan; and
how they relate to variability in behavior, cognition, and clinical outcomes.
Fine-grained brain functional organization#
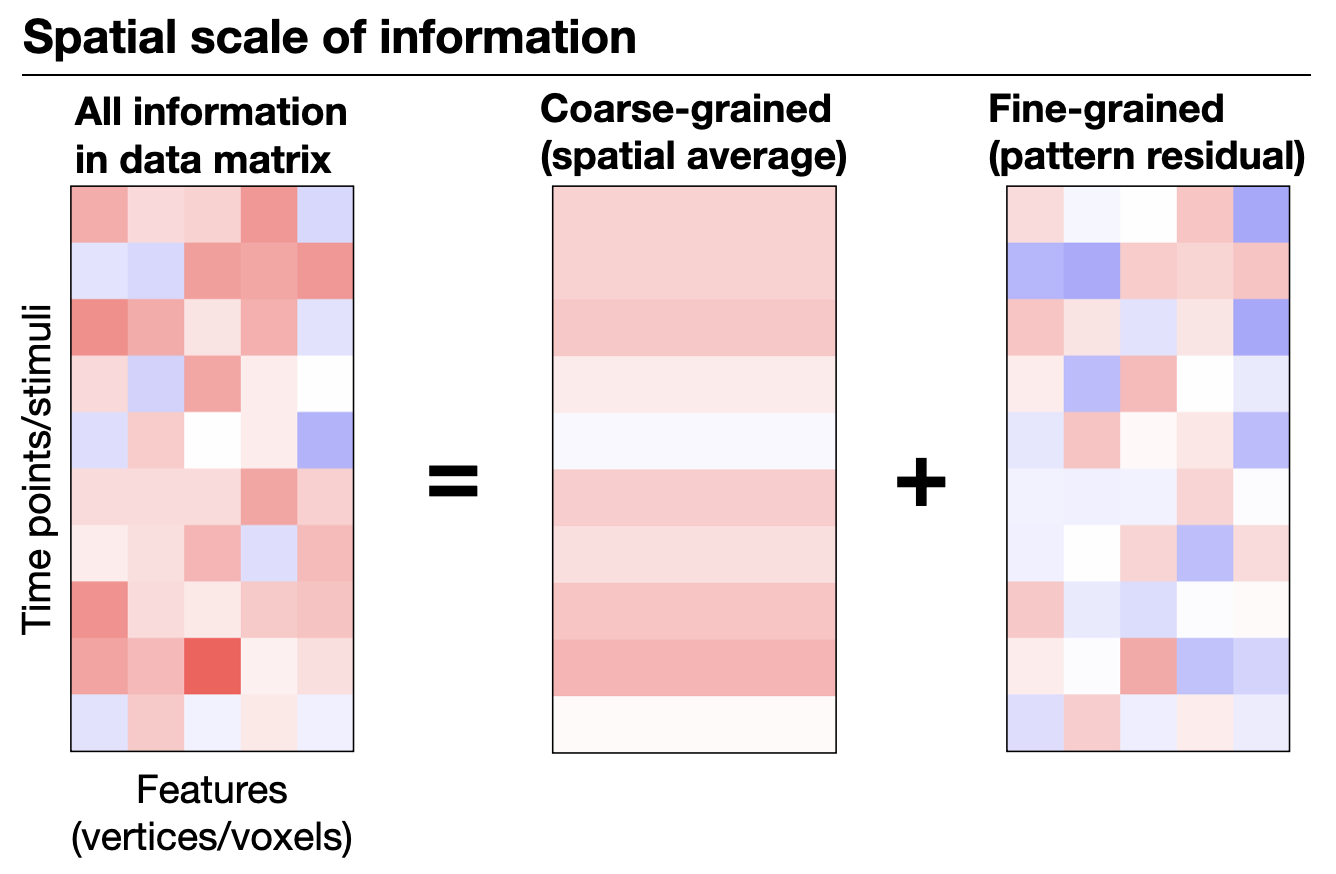
Fig 1. Coarse- vs. fine-grained information. Coarse-grained information is derived by spatially smoothing or averaging fMRI data within regions, reflecting their overall activity or connectivity (centimeter scale). Fine-grained information, in contrast, captures detailed vertex-by-vertex or voxel-by-voxel patterns within regions (millimeter scale).
We are particularly interested in individual differences in fine-grained brain functional organization — the detailed spatial patterns of brain activity and connectivity at the voxel or vertex level. Compared to coarse-grained differences (e.g., region-level averages), these fine-grained differences are
more reliable across independent data and more congruent across functional indices [Feilong et al., 2018];
more predictive of intelligence and other cognitive abilities [Feilong et al., 2021];
more experience-driven, potentially reflecting learning and training effects [Busch et al., 2024].
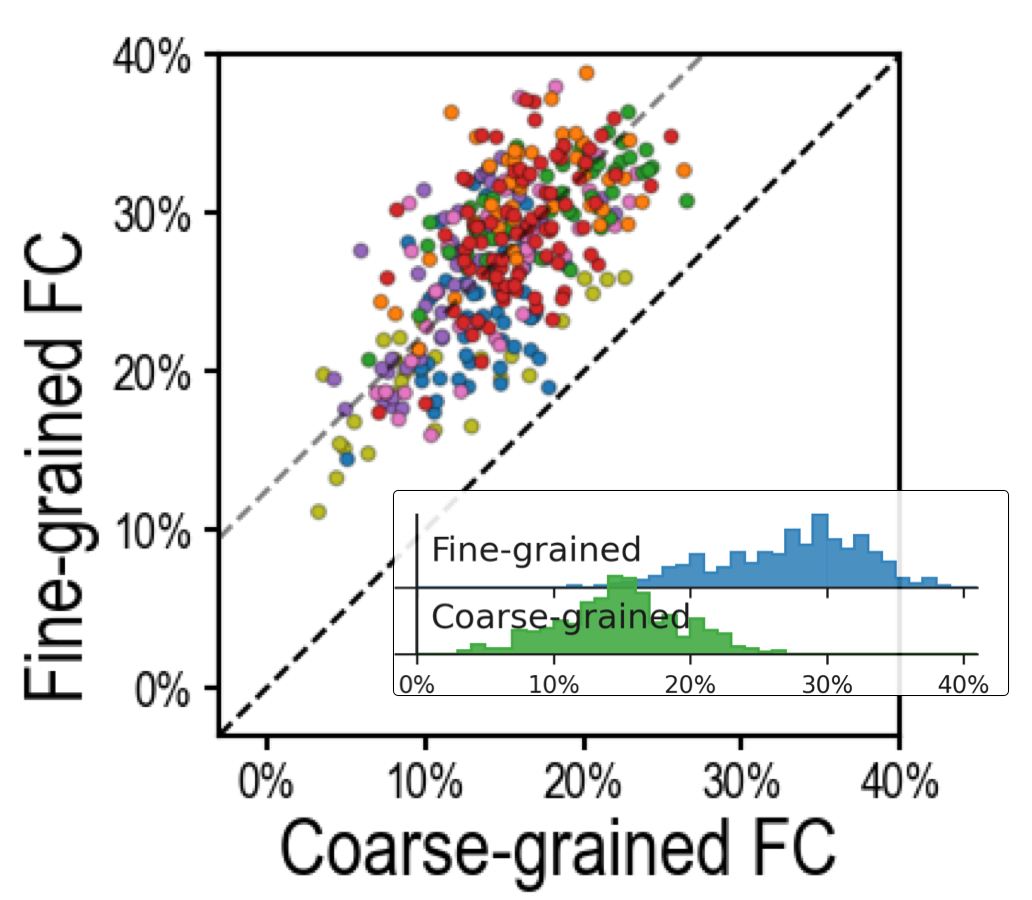
Fig 2. Predicting general intelligence (g). Fine-grained functional connectivity profiles (y-axis, blue histogram) are markedly better predictors of general intelligence than coarse-grained connectivity profiles (x-axis, green histogram) across different brain systems, accounting for twice as much variance in g. Adapted from Feilong et al. [2021, Figure 3]. Similar effects are observed for both task- and resting-state connectivity, and across different cognitive abilities [Feilong et al., 2021].
These fine-grained differences are often obscured by individual variability in anatomical–functional correspondence and cannot be properly studied using traditional alignment methods. We use hyperalignment [Haxby et al., 2020] and the Individualized Neural Tuning (INT) model [Feilong et al., 2023] to resolve these idiosyncrasies and reveal the individual differences in fine-grained functional organization.
References#
Ma Feilong, Samuel A Nastase, J Swaroop Guntupalli, and James V Haxby. Reliable individual differences in fine-grained cortical functional architecture. NeuroImage, 183:375–386, 2018. doi:https://doi.org/10.1016/j.neuroimage.2018.08.029.
Ma Feilong, J Swaroop Guntupalli, and James V Haxby. The neural basis of intelligence in fine-grained cortical topographies. elife, 10:e64058, 2021. doi:https://doi.org/10.7554/eLife.64058.
Erica L Busch, Kristina M Rapuano, Kevin M Anderson, Monica D Rosenberg, Richard Watts, BJ Casey, James V Haxby, and Ma Feilong. Dissociation of reliability, heritability, and predictivity in coarse-and fine-scale functional connectomes during development. Journal of Neuroscience, 2024. doi:https://doi.org/10.1523/JNEUROSCI.0735-23.2023.
James V Haxby, J Swaroop Guntupalli, Samuel A Nastase, and Ma Feilong. Hyperalignment: modeling shared information encoded in idiosyncratic cortical topographies. elife, 9:e56601, 2020. doi:https://doi.org/10.7554/eLife.56601.
Ma Feilong, Samuel A Nastase, Guo Jiahui, Yaroslav O Halchenko, M Ida Gobbini, and James V Haxby. The individualized neural tuning model: precise and generalizable cartography of functional architecture in individual brains. Imaging Neuroscience, 1:1–34, 2023. doi:https://doi.org/10.1162/imag_a_00032.
